A pull request with an implementation of forward feature selection waits in the Scikit-Learn repository since April 2017 (https://github.com/scikit-learn/scikit-learn/pull/8684), so we probably should get used to workarounds ;)
Table of Contents
To get an equivalent of forward feature selection in Scikit-Learn we need two things:
-
SelectFromModel class from feature_selection package.
-
An estimator which has either
coef_orfeature_importances_attribute after fitting.
Regression
In case of regression, we can implement forward feature selection using Lasso regression. This regression technique uses regularization which prevents the model from using too many features by minimalizing not only the error but also the value of the coefficients. It forces the model to set the coefficients of unimportant variables to 0 which means that such columns are not used.
import seaborn as sns
import pandas as pd
mpg = sns.load_dataset('mpg')
mpg = mpg.copy()
mpg = mpg.dropna()
# To simplify the code, I decided to drop the non-numeric columns
mpg.drop(columns = ['origin', 'name'], inplace=True)
from sklearn.feature_selection import SelectFromModel
from sklearn.linear_model import Lasso
X = mpg.drop(columns = ['mpg'])
y = mpg['mpg']
estimator = Lasso()
featureSelection = SelectFromModel(estimator)
featureSelection.fit(X, y)
selectedFeatures = featureSelection.transform(X)
selectedFeatures
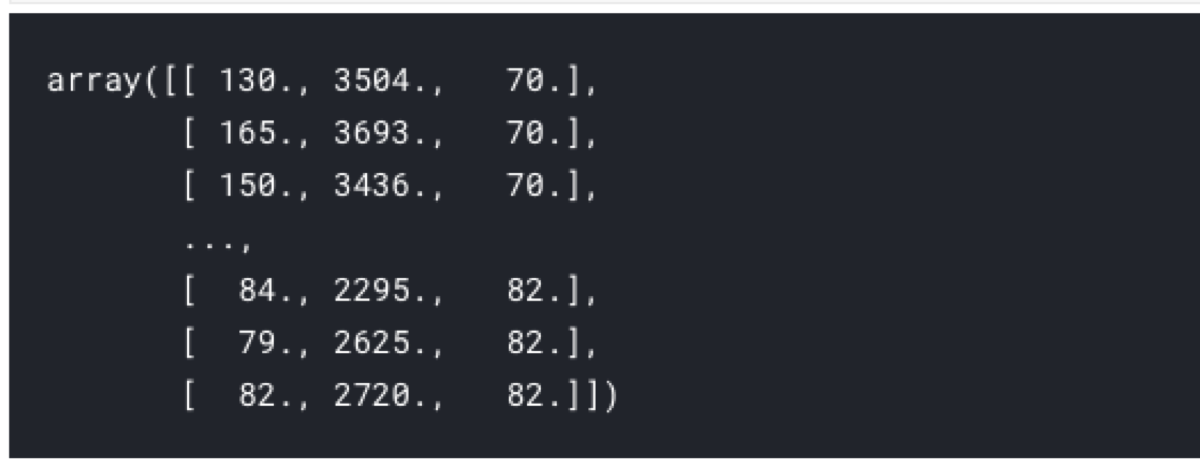
After fitting the model, we can filter the columns to get a list of variables used by the model.
X.columns[featureSelection.get_support()]

Classification
In the case of classification, forward feature selection may be replaced by fitting a decision tree-based classifier. ExtraTreesClassifier fits multiple decision trees. Every tree has access to a subset of given variables.
We can use either Gini impurity or information gain metric to select the feature which gives the best improvement of the classification result.
import seaborn as sns
import pandas as pd
titanic = sns.load_dataset('titanic')
titanic = titanic.copy()
titanic = titanic.dropna()
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import ExtraTreesClassifier
# There are some columns which contain the same value but have different names
X = titanic.drop(columns = ['survived', 'alive', 'class', 'who', 'adult_male', 'embark_town', 'alone'])
# Transform categorical variables to numeric values using one-hot encoding
X_one_hot_encoded = pd.get_dummies(X)
y = titanic['survived']
estimator = ExtraTreesClassifier(n_estimators = 10)
featureSelection = SelectFromModel(estimator)
featureSelection.fit(X_one_hot_encoded, y)
selectedFeatures = featureSelection.transform(X_one_hot_encoded)
selectedFeatures
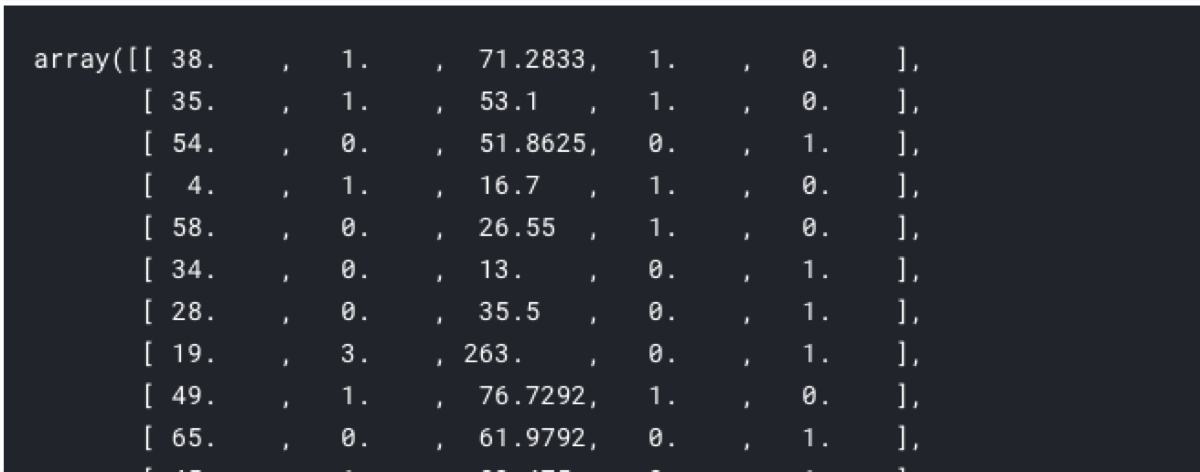
We can get the names of selected columns too.
X_one_hot_encoded.columns[featureSelection.get_support()]